Labs Recruiting Graduate Students
Labs Recruiting Graduate Students
Applications for graduate school are due December 1. All applicants must fulfill requirements of the MU Graduate School and show likelihood of successfully completing the departmental program. For more information about graduate admission to the Division of Biological Sciences, click on the button below.
The faculty listed below are currently recruiting graduate students to work on projects in their labs. Click on each faculty member's name to read about the lab and projects.
The Amato Lab is dedicated to uncovering the developmental origins of genital birth defects, which are among the most prevalent anomalies globally. Despite their frequency, the precise cause of these defects remain largely unknown and effective preventative measures are lacking. Using genetic mouse models, next-generation sequencing, organ culture techniques, and cutting-edge microscopy, the Amato lab aims to uncover the unknown causes of genital birth defects. Specifically, the lab focuses on identifying the function of unique cell types, the roles of hormone signaling, and the negative impacts of environmental exposures during the orchestration of genital formation. The goal is to then use the information gained from this research to develop pre-natal preventatives or surgical improvements for genital defects in humans.
Prospective students are encouraged to contact Dr. Ciro Amato to discuss potential projects.
Epithelia are the most common tissue type in animals. They can be found at organ boundaries, where they perform functions including absorption (the intestine), secretion (glands), filtration (kidneys), and gas exchange (the lungs). Epithelia are typically organized into sheets of cells that are one-cell thick, and our lab aims to understand how these sheets develop.
The Finegan Bergstralh Lab is currently working on two problems. 1) How do epithelial cells figure out the right direction in which to divide? 2) What happens when they don’t? These questions sit at the intersection of basic cell and developmental biology, biomedical research, and the physical sciences, so we take an interdisciplinary approach to answering them; our lab members come from several scientific backgrounds and use a range of techniques that includes traditional genetics, protein biochemistry, advanced light microscopy, and computational modelling.
Graduate student projects are tailored to suit student’s individual research interests, expertise, and career goals. We’d love to discuss opportunities for new lab members.
Prospective applicants should contact Dr. Bergstralh.
The Bhandari Lab is recruiting a student to work on CRISPR-dCas9 epigenome editing and epigenetic inheritance. Interested students are encouraged to contact Dr. Bhandari to discuss the project.
The lab of Dr. Jim Birchler focuses on genetics and genomics of maize with specific interests in developing synthetic chromosomes and new transformation techniques, the molecular basis of genomic balance, the molecular mechanism of the drive mechanism of the supernumerary B chromosome, and the molecular basis of heterosis, using maize as the model organism.
There are currently nine lab members consisting of technicians, postdocs, graduate students, and undergraduates. We have previously established foundational minichromosomes for genetic engineering and are currently developing procedures for gene stacking of individual genes and large segment additions on the minichromosomes. With regard to genomic balance, we are pursuing the molecular basis of why the addition or subtraction of a chromosome has a much more severe effect on an organism than changing the dosage of the whole set of chromosomes. These studies involve RNA-seq and chromatin analyses using the genetic tricks of maize. Our lab was involved in determining the sequence of the B chromosome of maize, which has properties to perpetuate itself despite that fact that it is a dispensable chromosome. CRISPR-Cas9 editing of candidate genes involved in this drive mechanism is being conducted to dissect their function in conjunction with genetics and cytological analyses. Heterosis is the increased biomass and vigor of hybrid plants, which has been used in agricultural practices to improve yield. We are using genetic and genomic tools to gain new insights into the molecular basis.
Students typically choose a project in which they are interested and are encouraged to develop it creatively within the context of the lab.
Watch video
Prospective applicants are encouraged to contact Dr. Jim Birchler to discuss potential projects.
Interested students are encouraged to contact Dr. David Braun to discuss projects.
Prospective applicants can reach Dr. Pamela Brown at brownpb(AT)missouri.edu.
Brown lab is a research community comprised of research technicians, undergraduate students, graduate students, and postdoctoral scholars. Dr. Brown is committed to investing in the success of her trainees and develops a customized mentoring plan for each individual. We welcome all individuals with an interest in microbial cell biology to learn more our research and lab environment.
Research overview: The Brown lab is interested in understanding the principles that govern bacterial morphology, a readily observable facet of microbial cell biology. Bacteria exhibit an amazing diversity of shapes and sizes that are precisely reproduced at every generation, indicating that morphology plays an important role in the life of these bacteria. Yet, we are still very far having a comprehensive understanding of how bacterial morphologies are generated. The Brown lab uses methods including bacterial genetics, quantitative cell biology, and microscopy to study growth patterning in the bacterial plant pathogen, Agrobacterium tumefaciens.
We invite you to meet some members of the lab and learn a little bit about our research projects by watching our LabBlitz 2020 video: https://youtu.be/KJOIBzMPIhg

Prospective applicants can reach Dr. Elizabeth Bryda at brydae@missouri.edu.
I am a molecular geneticist with >30 years of experience in the characterization of genes involved in human disease, the generation and genetic/phenotypic characterization of animal models and the development of various molecular tools for genetic testing of laboratory animals and cell lines. I am the Director of the Rat Resource and Research Center (RRRC), Director of the MU Animal Modeling Core (AMC) and co-Director of the Comparative Medicine Program. My research lab as well as the RRRC and AMC are located in a state-of-the art facility at the Discovery Ridge Research Park. Our team consists of a highly collaborative group that includes junior faculty, veterinarians, graduate students, undergraduates and research technicians. My lab develops and uses genetically modified rodent and zebrafish models to characterize genes and proteins relevant to a variety of diseases including hereditary deafness, polycystic kidney disease, inflammatory bowel disease, Alzheimer’s disease, and epilepsy. In addition, we study the impact of the microbiota and a variety of therapeutic agents/interventions including dietary/probiotic supplementation on disease phenotypes in animal models.
Examples of recent projects include making and characterizing green and red fluorescent rats, generating and characterizing rat models for the study of COVID-19, studying the effect of probiotic supplementation in zebrafish under stress and rats with Alzheimer’s Disease, analyzing the effects of dietary supplementation with plant-derived extracts on physiological measures in a rat model of obesity and diabetes, and developing methods for performing in vitro fertilization using fresh and frozen rat sperm.
Learn more about the Rat Resource and Research Center
Overview: The Chabu Lab is interested in understanding 1) how growth control signals are precisely integrated in developing tissues to generate mature organs with predictable size and 2) how some of these signals are corrupted in EGFR/RAS tumor tissues to promote robust tumor growth and drug resistance.
EGFR/RAS Tumor Growth and Drug Resistance: Oncogenic activation of the EGFR/RAS pathway activates a complex network of intracellular and intercellular signals that cause aggressive and drug resistant cancers. While we have made significant advances in our understanding of EGFR/RAS intrinsic signaling mechanisms, we know much less about intercellular EGFR/RAS signals and their role in cancer progression and drug resistance. Our laboratory and others have found that oncogenic EGFR/RAS activity triggers the release of extracellular vesicles (EV), which are known mediators of intercellular signaling. Further, it is known that EV mediate diverse pathologies, including nephropathies and cancers. Our most recent data using patients’ plasma EV and human cancer cells show that cancers-derived EV trigger mTOR/AKT activity, leading to resistance to EGFR-targeted therapies and chemotherapies. These findings and the current literature point to EV as potential targets for eliminating tumors and sensitizing patients to existing therapies. We have developed experimental tools and platforms to first understand the basic biology of EV-mediated cell-cell signaling (EV biogenesis, cargo loading, EV uptake in recipient cells, and the relevant cellular responses). These studies will potentially reveal a new class of molecular targets that can be exploited for therapeutic interventions.
Prospective applicants can reach Dr. Yves Chabu at chabuc(AT)missouri.edu.
Interested students are encouraged to contact Dr. Cornelison to discuss projects.
In the Cornelison Lab, we are interested in the cellular and molecular interactions that direct skeletal muscle development and regeneration both under normal conditions and during aging or disease. Our overarching question is, How do individual myogenic cells receive, process, and act on information from their surrounding environment in order to generate, maintain, and repair the ~650 unique skeletal muscles in the body, all of them the right size, in the right place, with the right connections to other tissues such as neural cells and blood vessels, so that the animal can move, grow, and survive?
During the past several years, we have focused primarily on the role of Eph:ephrin signaling in myogenesis. These juxtacrine signaling molecules are present in almost all cell types in vertebrates and can direct a wide variety of activities including survival, death, adhesion, repulsion, differentiation, dedifferentiation, transformation, and ‘stemness’. Our work has shown that Eph:ephrin interactions direct satellite cell motility (Stark et al., Development 2011), myofiber innervation (Stark et al., JCB 2015), and myocyte differentiation (Arnold et al., eLife 2020). Current projects funded through NIH include determining the transcriptional regulation of the interacting proteins ephrin-A5 and EphA7 in myoblasts and myocytes; characterizing the signaling complexes that ephrin-A5 and EphA7 are associated with on each cell; assessing the role of EphA7 in myocyte fusion; and testing whether inhibition of ephrin-A5 in vitro can enhance muscle stem cell specification for therapeutic cell transplant application. We use timelapse microscopy, immunohistochemistry and in situhybridization, cell culture, cell signaling assays, next-generation sequencing, and in vivo approaches in mouse to explore these questions.
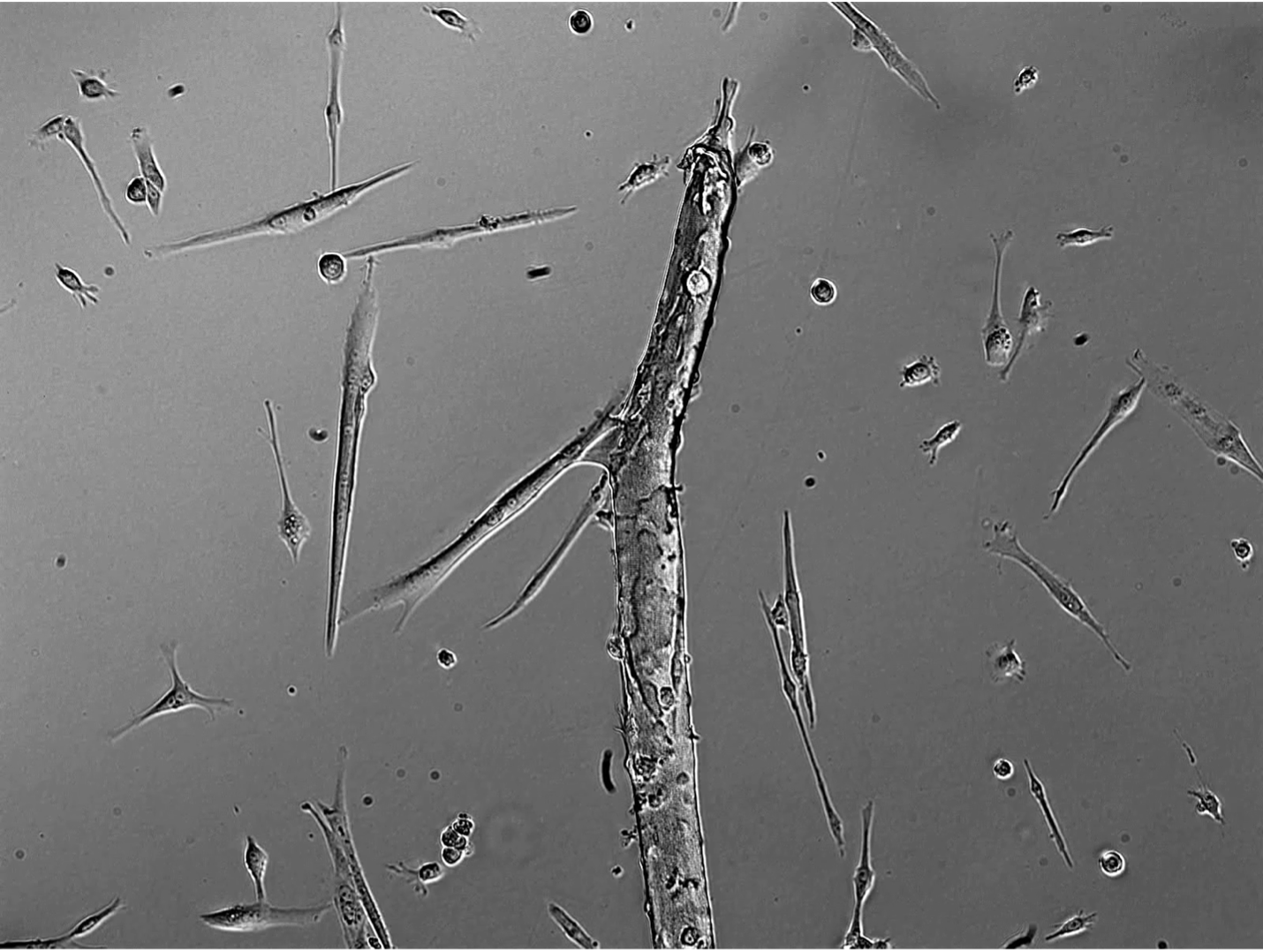
The lab currently has active collaborations with groups at MU working on computer vision, RNA aptamers, and volumetric muscle loss, as well as groups outside MU working on transcription factor binding and chromatin conformation and on novel transgenic tools to manipulate and study skeletal muscle regeneration. I encourage my graduate students and postdocs to collaborate broadly in order to grow their own network of colleagues and to bring outside expertise to bear on their chosen projects as appropriate.
The Goyes Lab is seeking motivated and curious graduate students to join our team in Fall 2026. Our lab investigates the reproductive ecology and behavior of amphibians, with a primary focus on the Emerald glass frog (Espadarana prosoblepon).
Students will have the opportunity to develop a project tailored to their interests and skillset, within the broader scope of the lab’s research program. Current and past projects include:
- Oviposition behavior and how environmental and substrate factors influence site selection
- Vocal behavior and communication strategies
- Mating systems and sexual selection
- Behavioral variation across habitats
- Future directions include the effects of anthropogenic change on reproductive behavior
We are also expanding into other systems, so applicants with interests in parental care, mating systems, and sexual selection are encouraged to reach out to Dr. Goyes Vallejos to discuss potential projects.
The Hamilton Lab is interested in the complex relationships between plants and the microbes that live in and around them, focusing on how these interactions can be either beneficial or harmful. Our studies focus on how the environment within a plant influences bacterial behavior, using the Ralstonia-tomato system to understand bacterial wilt disease and the Arabidopsis-Pseudomonas interaction to explore the difference between helpful and harmful bacteria. By examining the molecular mechanisms and communication signals that drive these relationships, the Hamilton Lab aims to uncover the factors that determine whether a microbe becomes a friend or foe and provide insights into our fundamental understanding of host-microbe interactions.
The Hamilton Lab is currently recruiting one gradaute student to join the lab for Fall 2026. Prospective students should contact Dr. Hamilton by email to discuss opportunities. The email should highlight how your background and goals align with those of the Hamilton Lab.
Prospective applicants can reach Dr. Casey Holliday at hollidayca(AT)missouri.edu.
The Holliday lab uses anatomical techniques, imaging and 3D biomechanical approaches to explore how animals are built, how they work and how they evolve. We tend to focus on the craniofacial biology and mechanics of reptiles including living species like crocs, birds and lizards, as well as their extinct ancestors like dinosaurs. We are experts in contrast-enhanced microCT, which allows us to see and measure soft tissues like muscles, brains, nerves and cartilages and are developing new methods in visualizing and quantifying complex anatomies of nerves, muscle architecture and joint systems in the skull and limbs, particularly in collaboration with Biology faculty member Kevin Middleton. We also keep a foot in the fossil record, using our understanding of living species to help guide our interpretations of how extinct species lived and how modern clades originated. We have shown how crocodilian skulls and feeding performance evolved, how blood vessels in dinosaur skulls aided in thermal biology, as well as how reconstructing cranial nerves help understand sensory biology in animals like Tyrannosaurus. We also like to migrate and exercise 3D models of anatomy into online and virtual reality environments.
Ongoing projects include:
1) 3D muscle architecture, comparative biomechanics and evolution of vertebrates.
We are in the throes of learning how accurately we can build 3D models of muscle fibers, how we can estimate physiological properties using these data, and how we can incorporate these data into developmental, biomechanical, ecological and evolutionary studies of jaw muscles and appendicular muscles. Notably, we are employing these methods to study the jaw muscles of birds and reptiles, their significance for the diversification of birds, patterns of ecomorphology in lineages of lizards and snakes, as well as in a growing sample of crocodilians and mammals. This work is all done in parallel with studies of skeletal morphology and mechanics so that we can build high fidelity 3D models of musculoskeletal systems to test a variety of evolutionary and ecological hypotheses. We are also using these approaches to study the origin of grasping in primates and other mammals via a collaborative grant with Carol Ward and Kevin Middleton thanks to the Leakey Foundation. Other ideas are welcome, the sky is the limit.
2) Biomechanics and evolution of crocodilians and dinosaurs (inc birds).
We have vested interests in learning how the heads of birds evolved from dinosaurs and how the heads of modern crocs evolved from their ancestors and all of the diversity in between. We spend time visiting museums, CT scanning fossils, and discovering how their joints are built, how their skulls work, and how these lineages have changed over the past 250million years. We’re currently trying to find traces of fossilized cartilage in joints with colleagues in Mizzou’s Geology Dept (Schiffbauer, Selly), understand how mandibular joints work in theropod dinosaurs, how the palate and cranial kinesis evolved along the line to birds and how patterns of somatosenstation and beaks evolved since the beginnings of the Mesozoic.
3) Evolution of mammalian jaw-ear musculature.
We know a lot about how the primitive jaw joint bones of mammal ancestors evolved into ear ossicles now used in hearing, but we know little about how the muscles evolved alongside the bones. We have an internal grant to explore this anatomy in growing sample of young and adult pigs, opossums and primates to better understand muscle-bone connections during the suckling-to-weaning transition with an eye towards getting at muscle evolution across mammals.
4) Biology of ‘exotic’ species.
We collaborate with a number of veterinarian colleagues which leads to a variety of projects that have proximal, medical outcomes, but also that feed into broader ecomorphological studies. Currently, along with several Avian veterinarians, we are using new contrast imaging approaches to study the sensory receptors inside the beaks of birds to help treat malocclusion and avoid aggressive beak trimming. This work will extend into studies of many other bird species, particularly parrots and ducks. We are also working on the anatomy of pangolins with colleagues from Chicago’s Brookfield Zoo.
We currently have 4 PhD students and 1 Undergraduate researcher in the lab. We try to find projects that fit with students’ passions and interests, but also play to our strengths. Everyone helps out with each other’s projects in a collaborative fashion. We work on crocodiles and dinosaurs, so we try our best to have fun and appreciate our jobs, while also doing some wicked science.

The Kang Laboratory is a part of the USDA Agricultural Research Service Biological Control of Insects Research Laboratory and is affiliated with the Division of Biological Sciences at the University of Missouri. We investigate the utilization of microbiomes and various genetic technologies in the control of insect pests that cause damage to American crops. Our interests are the environmental influences and within community interactions of microbiomes, host-microbe interactions, and their downstream impact on host immunity, metabolism, reproduction, and survival. We value teamwork, honest communication, and aim to foster an environment focused on the development and success of our students.
Our laboratory focuses on a multidisciplinary approach. First, we capture microbial and host variation in the wild. Next, we use statistical modeling to generate hypotheses. We then use the genetically tractable model organism Drosophila melanogaster to test and refine these hypotheses. The knowledge gained from these experiments is then utilized to control agricultural pests, such as the berry pest, Spotted Wing Drosophila (Drosophila suzukii). Current research efforts are described below. Our ultimate scientific goal is to combine these efforts to enable tailor-made microbial control of agricultural pests and enhance existing methods.
Microbial influence on Drosophila suzukii fitness.
Testing which microbes and what genes they possess are responsible for changes in the host.
Testing transcriptional changes made in the host by the microbes.
Microbial ecology of insect pests
Computational modeling of the wild Drosophila suzukii microbial assembly.
Testing microbial priority effects.
Utilizing artificial intelligence to identify metagenomic factors leading to the persistence of grape sour rot bacteria in the fly host
New genetic technologies towards the control of insect pests
RNAi pesticides (in collaboration with IU School of Medicine and the University of Notre Dame).
Creation of insect gene drive technology.
Computational modeling towards improved agriculture and food security
Machine-learned predictive models of viral infection risk.
Machine-learned habitat suitability maps of pest insects.
Interested students are encouraged to contact Dr. David Kang at Dave.Kang(AT)USDA.gov to discuss potential projects.
Prospective applicants can reach Dr. Elizabeth King at kingeg(AT)missouri.edu.
Research in the King Lab addresses fundamental questions in evolutionary genomics, seeking to understand how genomes change when phenotypes evolve. We integrate computational methods with large-scale empirical studies, with a primary focus on understanding the evolution of complex traits.
We are recruiting a Master’s student to work on an experimental evolution project in which we are selecting for flies that show high flight performance in an insect wind tunnel. This project is a collaboration with the Middleton lab and will focus on how flight-related phenotypes evolve in concert while also tracking the genomic changes that occur during adaptation. This project provides opportunities to learn image analysis, microscopy, experimental evolution, and genomic analysis.
Training new scientists (broadly defined) is one of the main goals of our lab, and we welcome prospective students interested in developing both their computational and empirical skills.
Saludos y gracias por su interés en unirse a nuestro equipo “Chipojo Lab”. I am a first-generation college student and have been extremely lucky to work with amazing graduate and undergraduate students that have contributed significantly to our research program.
I am a theory-driven behavioral ecologist and have established an integrative and interdisciplinary research program that reflects my interest in developing an intimate understanding of the behavior, ecology, and physiology of organisms under natural conditions, with the ultimate goal of elucidating evolutionary processes.
Research in the Chipojo Lab combines experimental and observational approaches, including long-term manipulations of natural populations. Our work is mostly done with tropical lizards in the genus Anolis. Our findings have greatly contributed to our knowledge of the evolutionary diversity of this group. More generally, our research has changed how we study communication and physiological constraints under natural conditions and how we view the behavioral capacity of reptiles.
Prospective applicants can contact Dr. Lei Lei at lln34(AT)health.missouri.edu.
Dr. Lei’s research focuses on ovarian reserve formation, maintenance, and its associated ovarian dysfunction and diseases. Oocytes have the unique ability to program early embryogenesis, but during a normal life cycle of a female, only a small number of mature oocytes (eggs) are produced. A drastic decline in egg quality in young/middle-aged women causes a significantly increased chance of infertility and birth defects. The loss of primary oocytes due to genetic mutations, aging and environmental and toxic factors leads to diminished ovarian function and an increased chance of ovarian cancer in postmenopausal women.
The goal of our research is to uncover the mechanisms of mammalian ovarian reserve formation and maintenance and to apply this fundamental knowledge to better understand female ovarian dysfunction and diseases, including premature ovarian insufficiency, infertility and ovarian cancer. Our previous studies revealed that during ovarian reserve formation in mouse fetal ovaries, oocytes differentiate via intercellular transport of organelle & cytoplasm among interconnected sister germ cells. In the differentiated oocyte, enriched organelles organize into a Balbiani body (B-body). The B-body is involved in ovarian reserve maintenance in adult ovaries through RNA storage and translational regulation. Currently, we are investigating in mice and humans: 1) establishment of fetal germ cell connectivity and its role in ovarian reserve formation; 2) how intercellular transport of organelles and cytoplasm during ovarian reserve formation influences ovarian function and female fertility in adulthood; 3) roles of organelle organization and RNA storage in ovarian reserve maintenance and activation in adult females; and 4) cellular origin and microenvironment of ovarian cancer. The long-term goal of our research is to decipher genetic and environmental factor-caused ovarian dysfunction and diseases and to develop novel medical solutions for early detection and prevention of ovarian diseases.
Dr. Mannie Liscum, liscumm@missouri.edu
All living organisms utilize both exogenous and endogenous cues to regulate their metabolism, growth and development. Plants, given their ‘fixed in the ground’ (sessile) nature are especially sensitive and responsive to their environment and exhibit highly plastic developmental programs. Unlike animals where the body plan is established almost entirely during embryogenesis, plant form and function is mostly elaborated post-embryonically. This dominance of post-embryonic patterning and growth affords plants the plasticity they need to modify their growth, morphology and behavior to even subtle and rapid changes in their environment. Because environmental inputs are numerous and ever changing, responsiveness of an organism is highly integrated, where multiple input signals are ‘sorted’ and transduced in an efficient manner to give rise to proper adaptive responses. Our laboratory is interested in how light acts as an environmental regulator of such integrated signal-response systems from molecules to functional ecology, and employ Arabidopsis thaliana as a model for most of our studies.
Though our laboratory has historically focused on phototropism as a model response, we have recently changed focus to examine the role of light in the modulation of reactive oxygen species (ROS) production and signaling. We are working to elucidate the molecule mechanisms by which phytochrome B (phyB), one of the five red/far-red photoreceptors in Arabidopsis, is integrated into the ROS responses to a variety of stress. Two elements of the ROS stress responses are of particular interest:
- How does cytoplasmic-localized phyB regulate the rapid production of ROS via plasma membrane-localized NADPH-oxidases (respiratory burst oxidase homolog, RBOH; termed NOX in animals)?
- How does nuclear-localized phyB regulate the long-term adaptation response that requires altered patterns of gene expression, and allows the plant to withstand exposure to subsequent stresses?
We are tackling these questions in collaboration with Profs. Ron Mittler (Division of Plant Sciences), Yosef Fitchman (Tel Aviv University), and Johanna Morrow (Westminster College; and Mizzou Biology PhD).
Prospective applicants can reach Dr. Paula McSteen at mcsteenp(AT)missouri.edu.
The McSteen lab uses a genetic approach to understand plant development. We start by screening for mutants in maize with defects in tassel and ear development and then identify the genes that are mutated and study how they work at the physiological, biochemical, cellular, genomic, genetic, molecular and even evolutionary level. Through this approach, we have identified genes involved in auxin biosynthesis, transport and response, nutrient uptake and stem cell signaling. Graduate student projects are available in multiple areas, all driven by our goal of understanding how meristems give rise to organs.
Project 1) The McSteen lab is best known for research on the plant growth hormone, auxin. See http://maizeauxre.missouri.edu/ for more information on our current NSF Plant Genome project. Our long-term goal is to understand how such a simple molecule as auxin has such diverse developmental effects in different tissues.
Project 2) The McSteen lab is currently funded by an internal grant to study the intersection between meristem development and environmental stress – in particular, nutrient deficiency. We have collaborative projects on the role of boron and iron in maize root development.
Project 3) The McSteen lab is just starting a collaborative project on vesicle trafficking in maize. We have identified multiple mutants defective in trafficking through their effects on auxin transport and plan to develop tools to study the role of vesicle trafficking in development, nutrient uptake and immunity.
Project 4) The McSteen lab also has an “EvoDevo” project on the evolution of inflorescence architecture in the grasses. Our graduate student on this project recently graduated so we have openings to pick up this project at a very exciting stage.
The lab is characterized by its team approach to maize genetics. Team McSteen is currently made up of a graduate student, a post-bac student and three undergraduate students. Check out some lab pictures at https://www.instagram.com/teammcsteen/

Research in our lab focuses on musculoskeletal form, ontogeny, and function in the contexts of animal feeding, locomotion, and evolution. We integrate studies across multiple scales from micro-scale structural materials of bone to macro-scale whole bone morphology to evolutionary changes across vertebrate taxa and with comparisons to extinct organisms. Recently, we are most often working on computational projects aimed at developing tools and techniques for visualizing 3D imaging and complex data sets.
Craniofacial Growth in Humans. For orthodontic and craniofacial surgical interventions, estimation of the timing of peak growth and/or the amount of remaining growth is important for planning treatment. We are seeking to estimate these quantities for a wide range of traits in two- and three-dimensional imaging. We collaborate with both basic scientists and clinicians with the overall goal of understanding the patterns of human craniofacial growth and variation. There are opportunities for developing methods for studying human growth, nonlinear modelling of growth, and 2D and 3D geometric morphometrics.
Evolution of the Flight System in Fruit Flies. We are recruiting a Master’s student to work on an experimental evolution project in which we are selecting for flies that show high flight performance in an insect wind tunnel. This project is a collaboration with the King Lab and will focus on how flight-related phenotypes evolve in concert while also tracking the genomic changes that occur during adaptation. This project provides opportunities to learn image analysis, geometric morphometrics, microscopy, experimental evolution, and genomic analysis.
Animals do not exist in a vacuum, and it is now widely recognized that an individual’s social environment can have substantial impacts on its fitness. Research in the Roth Lab integrates across multiple biological levels to explore the complex relationships between individuals and their social environment, as well as the consequences this has for ecology and evolution.
- Why are some individuals more social than others, and what are the costs and benefits of social connectivity?
- What drives social preferences between certain individuals, and how might this influence evolution?
- How can individual phenotypes predict group level traits (e.g., social network structure)?
- Moreover, how might these group level traits influence the fitness of individual group members, leading to feedback loops between individual phenotypes and group level traits?
The answers to such questions can help fill crucial gaps in our understanding of the evolutionary implications and selective consequences of animal sociality. Furthermore, we draw on sociality research to address fundamental questions in disease ecology, sexual selection, and animal personality (i.e., consistent between individual variation in behavior).
We use a variety of study systems for my work, with a focus on California quail (Callipepla californica) and three-spined stickleback (Gasterosteus aculeatus). Our work with quail uses radio frequency identification, coupled with blood collection and behavioral observations, to gain a deep understanding of the patterns and consequences (e.g., disease transmission, reproduction, sentinel dynamics) of the complex social behaviors exhibited by these fascinating birds. Our work with the stickleback utilizes a collaborative large-scale transplant experiment in Alaska to shed light on the social drivers of speciation.
Prospective graduate students are encouraged to contact Dr. Roth to discuss potential projects.
The Santin Lab is an integrative neurobiology lab broadly interested in how the brain generates healthy electrical activity, and in turn, how neural circuits acquire energy to support their synaptic and cellular functions. These interests fall into two main projects. First, we discovered that the frog brain shifts into a state that does not need oxygen and glucose to function. We are trying to understand the cellular, molecular, and metabolic mechanisms that allow neural circuits to reduce their energy demands but maintain seemingly normal function during conditions that resemble a stroke. Second, we study the neural processes that allow frogs to restart breathing after hibernating underwater for several months. Inactivity in neuromuscular systems usually leads to neurological issues. Thus, frog brains appear to have “figured out” how to avoid these problems. Although we work in comparative animal models, this work has implications for the discovery of novel treatments of stroke and neuronal injury, as we are uniquely positioned discover new forms of plasticity that improve fitness of neural circuits.
To answer these questions, we use a “genes to behavior” approach. This provides students with integrative training in diverse areas that include molecular biology, cellular neuroscience, and whole-animal physiology. We welcome students with experience in molecular methods and bioinformatics who wish to apply those skills to questions in neurophysiology. Likewise, we welcome students with a passion for animal physiology and behavior but would like to connect those processes to cellular and molecular mechanisms.
The Santin Lab is currently recruiting a graduate student with a background in -omics approaches and bioinformatics. Applicants are encouraged to contact Dr. Santin to discuss projects.
The Schul Lab studies the evolution of novel traits using the acoustic communication of Neoconocephalus katydids as model. Over the last 20 years, our work focused on the evolution of female preferences in response to rapidly diversifying male signals. While sensory work continues in the lab, our focus shifted over the last few years towards the evolution of novel call patterns: we aim to understand the genetics underlying changes of neural networks that result in new call patterns.
Males of Neoconocephalus triops have developmental plasticity of their calls: depending on the conditions experienced during maturation, two call morphs develop which differ for two call parameters at species defining levels; indeed, the two call morphs were initially described as different species. We can produce full siblings with different call morphs in the lab, indicating that these species level differences are due to gene expression. This allows us to study the divergence of these call traits integratively at behavioral, neurobiological, and transcriptome level within one species. This system will provide unique insights into the processes initiating the evolution of new signal traits and ultimately speciation. Other projects in the lab include aspects of sensory processing (e.g. the detection of change).These projects provide ample opportunity for dissertation projects that integrate multiple approaches and methods (e.g. neurophysiology, transcriptomics) and allow students to develop their on projects.
We are looking for one or two students who are excited about studying important evolutionary questions in an integrative way using this powerful system.
Prospective applicants can reach Dr. Johannes Schul at Schulz(AT)missouri.edu.
Interested students are encouraged to contact Dr. Laura Schulz to discuss projects.
In the orange bread mold Neurospora crassa, a gene present in one parent but not the other may originate from a selfish genetic element. Accordingly, this fungus maintains a genome defense mechanism known as meiotic silencing by unpaired DNA (MSUD), which silences unpaired genes (potential intruders) during sexual development. MSUD employs common RNA interference (RNAi) proteins, such as Argonaute and Dicer, to destroy target transcripts. The Shiu Lab investigates the genetic basis of this silencing mechanism. They have identified many genes involved in the MSUD pathway, and they utilize various tools to study the corresponding mutant phenotypes and protein functions (e.g., where the silencing factors go and what they may do).
Interested graduate students should contact Dr. Patrick Shiu to inquire about prospective projects.
Prospective applicants can reach Dr. Keith Slotkin at KSlotkin(AT)danforthcenter.org
We investigate how plant cells determine which fragments of DNA to “silence” the expression from. These regions of the genome that are commonly targeted for repression (“silencing”) are Transposable Elements (TEs), which are mobile ‘jumping genes’ that cause mutations when they jump. The overarching question driving our lab is how plant cells understand which regions of the genome are TEs to target silencing to.
In a broad sense, what the Slotkin laboratory aims to determine is how self-perpetuating feed-forward cycles in the cell are initiated. The cycles we work on are RNA interference (RNAi), RNA-directed DNA Methylation (RdDM) and maintenance epigenetic silencing. We have learned that these cycles all work to silence TEs, and can feed into each other. Our current goal is to determine how the first cycle of RNAi is triggered by TE transcripts.
Projects in the lab vary from proteomics to genomics, bench-work to purely computational. The majority of our research is done on the powerful reference plant Arabidopsis, while through collaborations we have also concentrated on maize, soybeans and aphids, and bioinformatically on many other genomes. Our expertise are in: Transposable Elements, Plant Epigenetics, RNAi, RdDM, small RNAs, DNA methylation and the Bioinformatics of multi-copy regions of the genome.
Visit the Slotkin Lab homepage at: https://slotkinlab.github.io
See what the lab has recently published: https://pubmed.ncbi.nlm.nih.gov/?term=slotkin+RK&sort=pubdate
Learn more about the Danforth Plant Science Center: https://www.danforthcenter.org
Interested students are encouraged to contact Dr. Van Goor to discuss projects.
Dr. Justin Van Goor is an evolutionary ecologist broadly interested in the maintenance and interactions of biological community associates, mostly using nematodes as a focal taxon. His research capitalizes on the fig-fig wasp mutualism as well as the multitude of other organisms (notably Parasitodiplogaster nematodes) that interact with these partners, as a model to better understand how communities are structured and are modulated over evolutionary timescales. His lab combines “classical” fieldwork, genomics/phylogenomics, greenhouse manipulations, and laboratory-based experiments.
Imagine a world where one could predict the phenotypes of a plant (or other organism) grown in any location around the world before ever testing it there. Imagine then that underneath those predictions were models that could teach you about the actual biology driving the phenotypes. Such predictions would revolutionize agricultural productivity, sustainability, climate change mitigation, conservation, and more! While we may never get to that level of accuracy and fidelity, each step in the iterative process of predicting and understanding phenotypes is as important as the final goal.
The Washburn Lab studies how plants interact with their genetics (G) and the environments (E) they are placed in. We use traditional phenotyping, drones, robots, physiological instruments, sequencing, and other methods to evaluate plants in the field, greenhouse, and growth chamber. We apply basic statistics, quantitative genetics, physiological modeling, machine learning, deep learning, and artificial intelligence to understand and predict biological, evolutionary, and agricultural systems. Current projects in the lab include: modeling GxE interactions in corn fields across the US, developing high-throughput phenotyping methods for mutant populations and germplasm collections, characterizing corn photosynthesis in cold weather and how corn might be planted earlier in the season, and determining how and why some corn lines use different biochemistries (C4 sub-types) for photosynthesis.
Prospective applicants can reach Dr. Jacob Washburn at Jacob.Washburn(AT)usda.gov.
Our lab seeks to understand why sexual reproduction and meiotic recombination are dominant in eukaryotic species despite their heavy evolutionary costs. To address this question, our research uses the microcrustacean Daphnia as a model system to investigate the causes and consequences of obligately asexual reproduction and the evolutionary forces driving the variation of meiotic recombination rate ad different biological scales (e.g., between populations/species). To achieve these goals, our lab currently uses a combination of high-throughput genomic and single-cell sequencing, bioinformatic genomic analysis, and CRISPR-Cas gene editing. We are looking for multiple graduate students interested in participating in the following research areas.
The genetic mechanisms underlying the origin of obligate parthenogenesis in Daphnia.
In this project, we are interested in exploring the genetic mechanisms giving rise to obligately parthenogenetic Daphnia. Our previous studies strongly suggest that the genetic incompatibility between two sexual species D. pulex and D. pulicaria is critical for the origin of obligate parthenogenesis in the hybrids of these two species.This project would be highly interesting for students who would like to combine evolutionary genomic analysis with cellular and molecular biology work to test the roles of specific genes and genetic variants in the origin of obligate parthenogenesis.
The evolution and genetic basis of meiotic recombination rate variation in Daphnia.
In this project, we are investigating the genetic variants regulating meiotic recombination rate variation across Daphnia populations/species and are examining whether meiotic recombination evolves in an adaptive fashion. To estimate recombination rate and identify variants associated with recombination rate variation, we are mainly using and developing single-sperm whole-genome sequencing methodologies and population genomic analysis. We will also use CRISPR-Cas genetic editing to modify genetic variants to examine their effect on recombination rate. This project is ideal for someone who is interested in combining their passion in evolution, single-cell sequencing, and genetic editing in one project.
CRISPR-Cas genetic editing in Daphnia.
Our lab is actively developing CRISPR-Cas gene editing methodologies for Daphnia, currently including gene knock-out, gene knock-in, and precise base editing. We have a goal of creating 1000 Daphnia mutants in the next 4-5 years and test their gene knock-out effects to gain a deeper understanding of gene function and interactions. For this research direction, we are looking for students with passion in mastering CRISPR gene editing and understanding gene functions and interactions.
How environmental mutagens affect genomic integrity.
We are interested in understanding the effect of different environmental mutagens, for example, ethyl methanesulfonate (EMS), on the mutation rate and spectrum and gene expression in Daphnia. We use mutant genomic analysis with gene knock-out mutants to understand how different genes contribute to safeguarding genome integrity against the harmful effect of mutagens. This project is desirable for students with passion in gaining a deep understanding of the interaction of environment and genomes.
Interested students are encouraged to contact Dr. Bing Zhang to discuss projects.
The Zhu Lab leverages advanced single-cell/spatial transcriptomics combined with confocal microscopy to dissect the spatial organization of plant-microbe interactions at cellular and molecular levels. The research aims to deepen our understanding of the spatial dynamics of plant-microbe interactions and provide insights into developing disease-resilient plants.
Prospective students are encouraged to contact Dr. Zhu to discuss potential projects.
Dr. Manuel Leal
Director of Graduate Studies
lealm@missouri.edu















