New tool to visualize whole chromosomes in plants
A team of scientists led by MU maize geneticist James Birchler has designed a tool for scientists to paint whole chromosomes in plants different colors. The method is described in a paper published this month in the Proceedings of the National Academy of Sciences.
“The ability to visualize whole chromosomes in different colors is an extremely valuable tool for studying how chromosomes work,” said Birchler, who is a Curators’ Distinguished Professor in the Division of Biological Sciences. “This type of technique has been successfully used and applied to study human chromosomes. For some time, we have been interested in developing a way that this could be used in the plant world. This paper is the foundation for how this can be done.”
The most common tool scientists use to visualize chromosomes or parts of chromosomes is called fluorescence in situ hybridization, or FISH. FISH uses a piece of DNA, called a probe, that is labeled with a fluorescent molecule to “paint” each chromosome pair a different color. The technique relies on each probe to be complementary to a unique region of one chromosome. The problem with plants is an overabundance of repetitive DNA, or similar looking sequences that appear on multiple chromosomes, that interferes with the specificity of the tool. The new method overcomes this problem by deleting all repetitive sequences during the design process, leaving only those stretches of DNA specific to individual chromosome pairs to be used as probes.
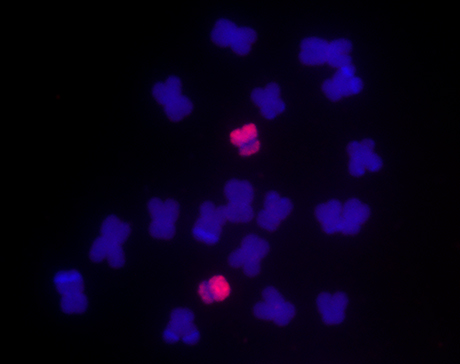
The researchers developed paints for all ten chromosome pairs in maize and, as proof of concept, applied each chromosome-specific paint to chromosomes isolated from root tip cells. Here, chromosome 10 is painted in red, showing the specificity of the tool.
“The idea to use a reference genome to bioinformatically identify short sequences specific to each chromosome pair is the real breakthrough,” said Birchler. “The bioinformatics procedure is an improved way of going about making the chromosome collections and represents an advance over the way it’s normally done, even in humans, because these probes can be amplified and reused over and over again.”
The lab of Jiming Jiang at Michigan State University led the bioinformatic work and generated the sequences. Birchler and his team used the sequences to develop FISH paints for each of the ten chromosome pairs in maize (corn) and, as proof of concept, applied each chromosome-specific probe to chromosomes isolated from maize root tips, where dividing cells are common. They also applied selected chromosomal paints to a variety of known chromosomal aberrations as well as chromosomes at different stages of the cell cycle.
“One of the really exciting aspects of this technology is that it will allow one to follow a single chromosome pair through the process of meiosis. It will also allow one to see how individual chromosomes are organized in the interphase nucleus. If we combine this technique with single gene FISH, then we will be able to see how individual chromosomes and individual genes are behaving in the interphase nucleus, which was previously a black box,” he said.
The researchers made the chromosome-specific sequences for maize publicly available. While the tool was worked out in maize, the researchers said that analogous probe libraries can be developed in any species, plant or otherwise, with a reference genome.
“With whole genome sequences and the ability to bioinformatically pick out short sequences as probes, we now have a more direct approach for making a probe collection. That’s an important advance over previous chromosome painting techniques,” said co-first author Patrice Albert. Albert, who is a senior research specialist in the Division of Biological Sciences, made the probes and conducted FISH experiments.
Birchler said the new tool is useful to scientists studying chromosomes and effects of chromosomal aberrations, such as missing or extra pieces, or parts from different chromosomes that have swapped places. The insights gained with the tool however may have broad application.
“Understanding the basics of how chromosomes work, as for any kind of basic research, very often leads to valuable, applied results usually not originally anticipated but eventually highly valuable,” he said. “Once one has these tools to learn new things, then the whole community becomes aware of it. Then the whole community is thinking about how this can be applied. That’s valuable for the eventual impact it may have for practical applications.”
Albert, P.S., Zhang, T., Semrau, K., Rouillard, J.-M., Kao, Y.-H., Wang, C.-J.R., Danilova, T.V., Jiang, J., Birchler, J.A. Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships (2019) Proceedings of the National Academy of Sciences of the United States of America, 116 (5), pp. 1679-1685.